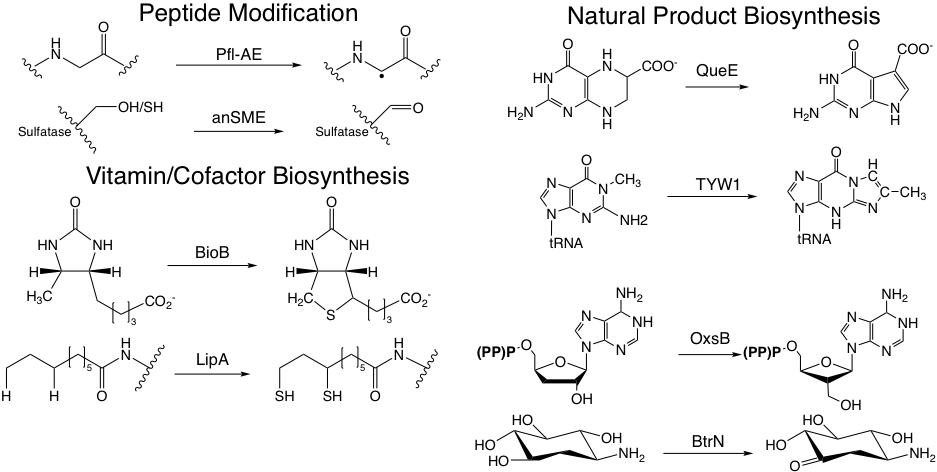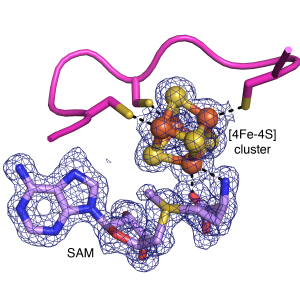
The 100,000-membered radical S-adenosylmethionine (SAM) enzyme superfamily is a Drennan lab favorite in their quest to understand how Nature uses radical cofactors to perform the most challenging of chemical transformations. Radical SAM enzymes perform a range of functions from vitamin and antibiotic biosynthesis to post-translational modifications (some reactions of radical SAM enzymes are shown in the accompanying figure). The Drennan lab started their work on radical SAM enzymes in collaboration with the Jarrett laboratory (U Hawaii) to investigate the structural basis of sulfur insertion by biotin synthase. The biotin synthase structure was one of the first two X-ray structures of a radical SAM enzyme, which established the ‘core’ fold for this enormous superfamily. More recently, Drennan (in collaboration with Squire Booker at Penn State U) solved a structure of lipoyl synthase that shows an Fe4S4 cluster being cannibalized for its sulfur. Using the Laboratory for Anaerobic Crystallography (LAC@MIT), they also structurally interrogate radical SAM enzymes that contain additional iron-sulfur clusters (with Booker and Vahe Bandarian (U Utah)) or cobalamin (with Hung-wen Liu, U Texas-Austin). The functions of these enzymes include peptide and nucleic acid modification, and biosynthesis of antibiotic and antiviral compounds. We also study glycyl radical enzyme (GRE) activases, the radical SAM enzymes that post-translationally modify glycine to form an enzyme-bound glycyl radical species.
Press
Chemical and Engineering News (2021): “Structure Reveals Why Radical Enzyme isn’t Radical” by Celia Henry Arnaud. Vol. 99, Issue 4.
Nat. Chem. Biol. (2017): “Radical Ring Resizing” by Caitlin Deane vol. 13, 569.
MIT News August 8, 2016, MITEI News and Office of Science homepage as a University Research Highlight: “Research by MIT Undergrad Helps Crack Chemical Mystery” by Peter Dizikes.
Publications (‡Co-first authors, *Corresponding authors)
Andorfer, M.C., King-Roberts, D.T., Imrich, C.N., Brotheridge, B.G., and Drennan*, C.L. (2023) Development of an in vitro method for activation of X-succinate synthases for fumarate hydroalkylation, iScience 26(6), 106902. doi: 10.1016/j.isci.2023.106902. PMCID: PMC10239695. PMID: 37283811
Knox, H.L., Chen, P.Y.-T., Blaszczyk, A.J., Mukherjee, A., Grove, T.L., Schwalm, E.L., Wang, B., Drennan*, C.L., and Booker*, S.J. (2021) Structural Basis for Non-Radical Catalysis by TsrM, a Radical SAM Methylase, Nat. Chem. Biol. online ahead of print. doi: 10.1038/s41589-020-00717-y
Grell, T.A.G., Bell, B.N., Ngyuen, C., Dowling, D.P., Bruender, N.A., Bandarian, V., and Drennan*, C.L. (2018) Crystal Structure of AdoMet Radical Enzyme 7-Carboxy-7-Deazaguanine Synthase from Escherichia coli Suggests How Modifications Near [4Fe-4S] Cluster Engender Flavodoxin Specificity, Protein Science 28, 202-215. PMCID: PMC6295903.
Grell, T.A.G., Kincannon, W.M., Bruender, N.A., Blaesi, E.J., Krebs*, C., Bandarian*, V., and Drennan*, C.L. (2018) Structural and Spectroscopic Analysis of Sporulation Killing Factor Biosynthetic Enzyme SkfB, an AdoMet Radical Sactisynthase, J. Biol. Chem. 293, 17349-17361. PMCID: PMC6231123
Grell‡, T.A.J., Young‡, A.P., Drennan*, C.L., and Bandarian*, V. (2018) Biochemical and Structural Characterization of a Schiff Base in the Radical-Mediated Biosynthesis of 4-Demethylwyosine by TYW1, J. Am. Chem. Soc. 140, 6842-6852. PMCID: PMC5994729 Faculty of 1000 Recommended
Bridwell-Rabb‡, J., Zhong‡, A., Sun, H.G., Drennan*, C.L., and Liu*, H.-w. (2017) A B12-Dependent Radical SAM Enzyme Involved in Oxetanocin A Biosynthesis, Nature 544, 322-326. PMCID: PMC5398914 Faculty of 1000 Recommended
Shisler, K.A., Hutcheson, R.U., Horitani, M., Duschene, K.S., Crain, A.V., Byer, A.S., Shepard, E.M., Rasmussen, A., Yang, J., Broderick, W.E., Vey, J.L., Drennan, C.L., Hoffman, B.M., and Broderick*, J.B. (2017) Monovalent Cation Activation of the Radical SAM Enzyme Pyruvate Formate-Lyase Activating Enzyme, J. Am. Chem. Soc. 139, 11803-11813. PMCID: PMC5579537
Bruender, N.A., Grell, T.A.J., Dowling, D.P., McCarty, R.M., Drennan, C.L., and Bandarian*, V. (2017) 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-L-methionine Enzyme with Polar Tendencies, J. Am. Chem. Soc. 139, 1912-1920. PMCID: PMC5301278
McLaughlin, M.I., Lanz, N.D., Goldman, P.J., Lee, K.-H., Booker, S.J., Drennan*, C.L. (2016) Crystallographic Snapshots of Sulfur Insertion by Lipoyl Synthase, Proc. Natl. Acad. Sci. U.S.A. 113, 9446-9450. PMCID: PMC5003258
Dowling, D.P., Bruender, N.A., Young, A.P., McCarty, R.M., Bandarian, V., Drennan*, C.L. (2014) Radical SAM Enzyme QueE Defines a New Minimal Core Fold and Metal-dependent Mechanism, Nat. Chem. Biol. 10, 106-112. PMCID: PMC3939041 Faculty of 1000 Recommended
Goldman, P.J., Grove, T.L., Sites, L.A., McLaughlin, M.I., Booker, S.J., Drennan*, C.L. (2013) X-ray Structure of an AdoMet Radical Activase Reveals an Anaerobic Solution for Formylglycine Posttranslational Modification, Proc. Natl. Acad. Sci. U.S.A. 110, 8519-8524. PMCID: PMC3666706
Goldman, P.J., Grove, T.L., Booker, S.J., and Drennan*, C.L. (2013) X-ray Analysis of Butirosin Biosynthetic Enzyme BtrN Redefines Structural Motifs for AdoMet Radical Chemistry, Proc. Natl. Acad. Sci. U.S.A. 110, 15949-15954. PMCID: PMC3791736
Vey, J.L., Yang, J., Li M., Broderick, W.E., Broderick, J.B., and Drennan*, C.L. (2008) Structural Basis for Glycyl Radical Formation by Pyruvate Formate-Lyase Activating Enzyme, Proc. Natl. Acad. Sci. U.S.A. 105, 16137-16141. PMCID: PMC2571006
Berkovitch, F., Nicolet, Y., Wan, J.T., Jarrett, J.T., and Drennan*, C.L. (2004) Crystal Structure of Biotin Synthase, an S-Adenosylmethionine-Dependent Radical Enzyme, Science 303, 76–79. Faculty of 1000 Recommended
Reviews
Ulrich, E.C. and Drennan*, C.L. (2022) The Atypical Cobalamin-Dependent S-Adenosyl-L-Methionine Nonradical Methylase TsrM and its Radical Counterparts, J. Am. Chem. Soc. 144(13), 5673-5684. https://doi.org/10.1021/jacs.1c12064. PMCID: PMC8992657. PMID: 35344653
Bridwell-Rabb*, J., Li, B., Drennan, C.L. (2022) Cobalamin-dependent Radical S-adenosylmethionine Enzymes: Capitalizing on Old Motifs for New Functions, ACS Bio. & Med. Chem. 2(3), 173-186. doi: 10.1021/acsbiomedchemau.1c00051. PMCID: PMC9204698
Bridwell-Rabb*, J., Grell, T.A.J., and Drennan*, C.L. (2018) A Rich Man, Poor Man Story of S-adenosylmethionine and Cobalamin Revisited, Ann. Rev. of Biochem. 87, 555-584. PMC Exempt – Invited Review. DOI: 10.1146/annurev-biochem-062917-012500
Grell, T.A.J., Goldman, P.J., and Drennan*, C.L. (2015) SPASM and Twitch Domains in S-Adenosylmethionine (SAM) Radical Enzymes, J. Biol. Chem. 290, 3964-3971. PMCID: PMC4326806
Dowling, D.P., Vey, J.L., Croft, A.K., and Drennan*, C.L. (2012) Structural Diversity in the AdoMet Radical Enzyme Superfamily, Biochim Biophys Acta 1824, 1178-1195. PMCID: PMC3523193
Vey, J.L., and Drennan*, C.L. (2011) Structural Insights into Radical Generation by the Radical SAM Superfamily, Chem. Rev. 111, 2487–2506. PMC Exempt – Invited Review. DOI: 10.1021/cr9002616