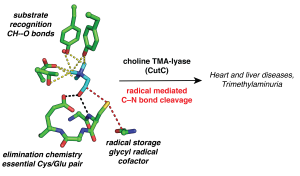
Glycyl radical enzymes (GREs) are abundant in the human gut microbiome. They use a post-translationally generated enzyme-bound glycyl radical species to carry out radical-based chemistry in anoxic environments. Our collaborator, Professor Emily Balskus (Harvard University), has pioneered a profiling strategy to discover novel GREs that are enriched in the human microbiome, and together, we carry out structure/function analyses of the resulting GREs. We recently solved a series of structures of choline trimethylamine-lyase (CutC), which produces trimethylamine (TMA), a microbial metabolite that is associated with nonalcoholic fatty acid liver disease, atherosclerosis, heart disease, diabetes and the metabolic disorder trimethylaminuria (fish malodor syndrome). We are currently working with the Balskus lab on inhibitor design in an attempt to provide the first medical remedy for fish malodor syndrome and to probe the importance of CutC activity to microbial communities and its impact on host health. We are also working on structurally characterizing newly discovered GREs such as hydroxy-L-proline dehydratase and probing mechanisms for GRE repair.
Press
MIT Biology News Briefs Mar 26, 2021, “Spying on enzymes while they perform chemical reactions could help treat gut ailments” by Raleigh McElvery.
MIT News Mar 17, 2020, “Bacterial enzyme could become a new target for antibiotics” by Anne Trafton.
MIT Biology News Briefs Jul 17, 2019, “Bacteria use ‘spare part’ proteins to repair damage and survive inhospitable conditions” by Saime Sidik.
Publications
Andorfer, M.C., King-Roberts, D.T., Imrich, C.N., Brotheridge, B.G., and Drennan*, C.L. (2023) Development of an in vitro method for activation of X-succinate synthases for fumarate hydroalkylation, iScience 26(6), 106902. doi: 10.1016/j.isci.2023.106902. PMCID: PMC10239695. PMID: 37283811
Andorfer, M.C., Backman, L.R.F., Li, P.L., Ulrich, E.C., Drennan, C.L., (2021) Rescuing activity of oxygen-damaged pyruvate formate-lyase by a spare part protein. J. Biol. Chem. Online Now. doi: 10.1016/j.jbc.2021.101423
Dawson, C.D., Irwin, S.M., Backman, L.R.F., Le, C., Wang, J.X., Vennelakanti, V., Yang, Z., Kulik*, H.J., Drennan*, C.L., Balskus*, E.P. (2021) Molecular Basis of C-S Bond Cleavage in the Glycyl Radical Enzyme Isethionate Sulfite-Lyase. Cell Chem. Biol. Online Now. doi: 10.1016/j.chembiol.2021.03.001
Bollenbach, M., Ortega, M., Orman, M., Drennan, C.L., and Balskus, E.P. (2020) Discovery of a Cyclic Choline Analog that Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria, ACS Med. Chem. Lett. 11, 1980-1985. PMCID: PMC7549264.
Backman‡, L.R.F., Huang‡, Y.Y., Andorfer, M.C., Gold, B., Raines, R.T., Balskus*, E.P., and Drennan*, C.L. (2020) Molecular Basis for Catabolism of the Abundant Metabolite Trans-4-hydroxy-L-Proline by a Microbial Glycyl Radical Enzyme, eLife 9, e51420 PMCID: PMC7077986.
Bowman, S.E.J, Backman, L.R.F., Bjork, R.E., Andorfer, M.C., Yori, S., Caruso, A., Stultz, C.M., and Drennan*, C.L. (2019) Solution Structure and Biochemical Characterization of a Spare Part Protein That Restores Activity to an Oxygen-damaged Glycyl Radical Enzyme, J. Biol. Inorg. Chem. Special Issue in Honor of Joan Broderick 24, 817-829. PMCID: PMC6754787.
Orman, M., Bodea, S., Funk, M.A., Martinez-del Campo, A., Bollenbach, M., Drennan, C.L., and Balskus*, E.P. (2018) Structure-Guided Identification of a Small Molecule that Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria. J. Am. Chem. Soc. 141, 33-37. PMCID: PMC6475491.
Bodea‡, S., Funk‡, M.A., Balskus*, E.P., and Drennan*, C.L. (2016) Molecular Basis of C–N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase, Cell Chem. Biol. 23, 1206-1216. PMCID: PMC5493019
Review Article
Backman, L.R.F., Funk, M.A., Dawson, C.D., and Drennan*, C.L. (2017) New Tricks for the Glycyl Radical Enzyme Family, Crit. Rev. Biochem. Mol. Biol. 52, 674-695. PMCID: PMC5911432